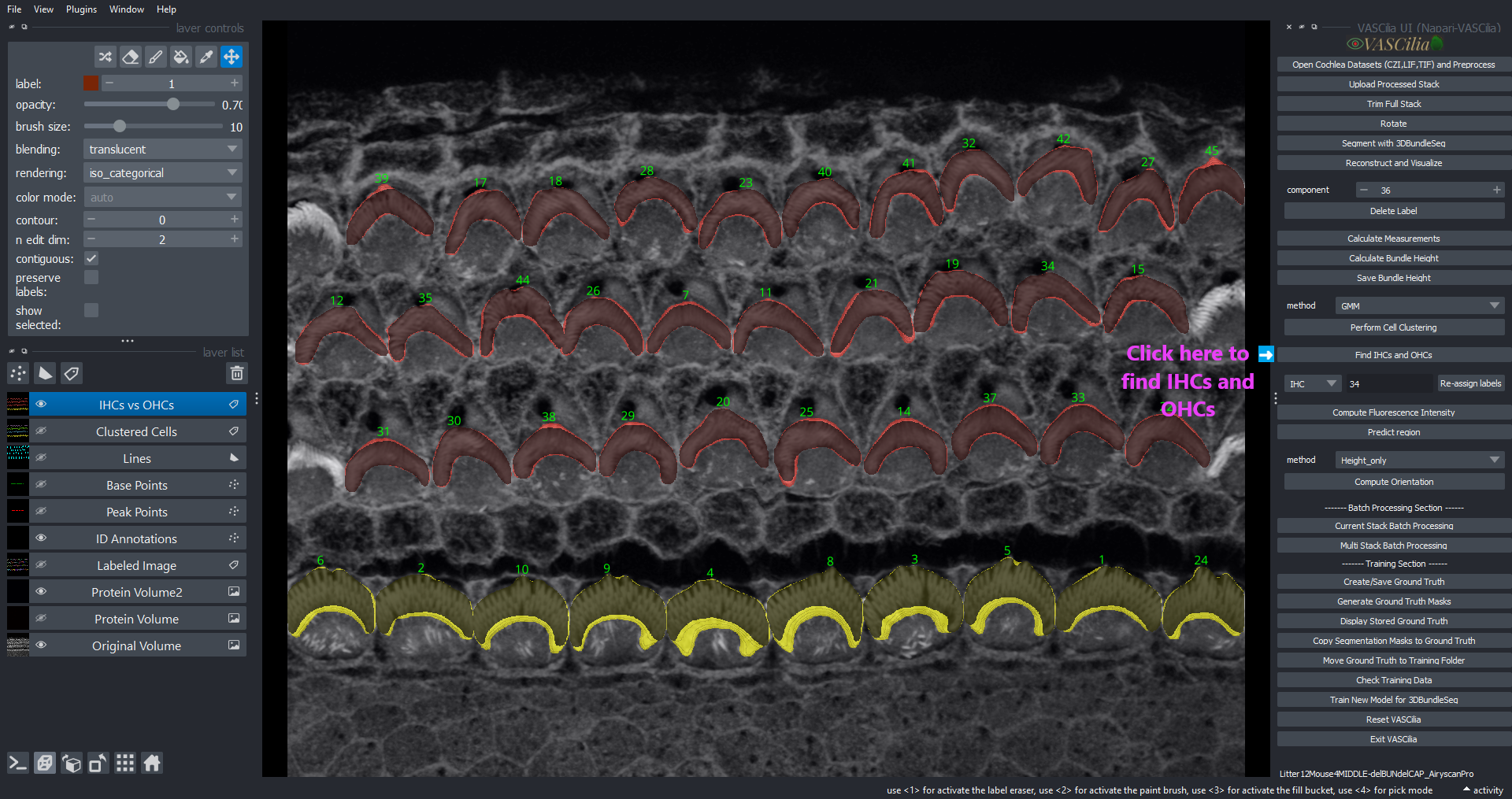
Cell Clustering#
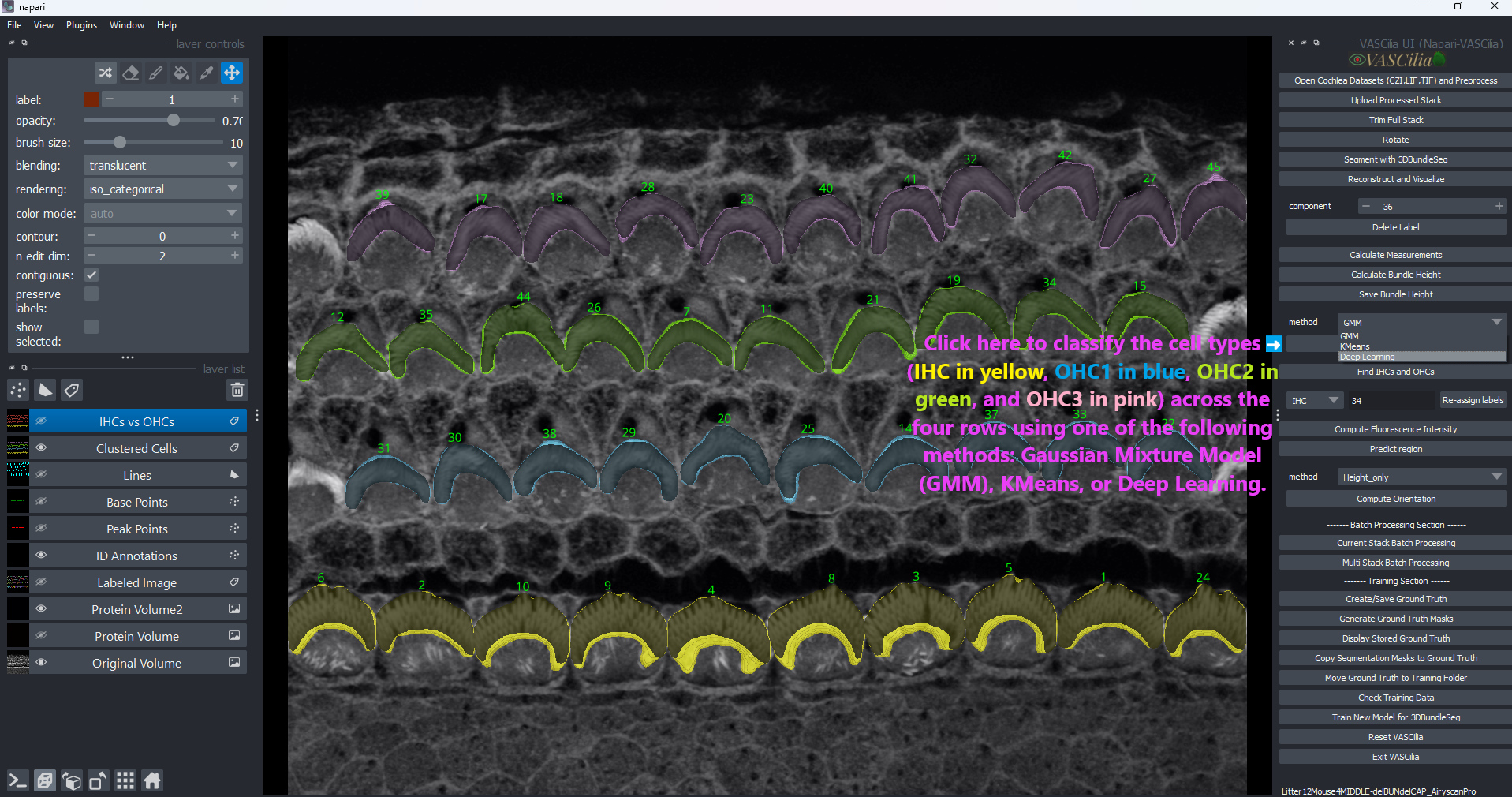
The Cell Clustering feature in VASCilia enables the classification of cochlear cells into specific types such as Inner Hair Cells (IHCs) and Outer Hair Cells (OHCs) using various clustering methods. This feature supports both unsupervised clustering techniques and deep learning models for accurate cell identification.
Key Features#
Clustering Methods:
Gaussian Mixture Models (GMM): Clusters cells based on Gaussian distribution assumptions.
KMeans: Groups cells using distance-based partitioning.
Deep Learning: Leverages pretrained deep learning models for advanced cell classification.
Cell Types:
Inner Hair Cells (IHC).
Outer Hair Cells (OHC), including subtypes: - OHC1 - OHC2 - OHC3
Interactive Reassignment: - Allows users to manually reassign cell labels to specific types using an intuitive interface.
Visualization: - Displays clustered cells in distinct layers:
Clustered Cells: Colored visualization of all clusters.
IHCs vs OHCs: Separate visualization for IHCs and OHCs.
Distance Integration: - Updates the saved physical distances with corresponding cell type classifications.
Workflow#
### Perform Cell Clustering
Choose a clustering method:
GMM or KMeans: Groups cells based on centroid positions.
Deep Learning: Uses signal intensity and pretrained models to classify cells.
Click Perform Cell Clustering to initiate the clustering process.
Review the resulting clustered layers.
### Reassign Clustering
Select the desired cell type (e.g., IHC, OHC1).
Enter the label number to reassign.
Click Re-assign Labels to update the clustering.
Practical Considerations#
Clustering Preconditions:
Ensure that bundle height calculation is complete and saved before performing clustering.
Deep Learning Models:
Utilizes pretrained models (best_model_ihc_v3.pth, etc.) for precise cell classification.
Visualization:
Clustered Cells: Uses distinct colors for easy identification.
IHCs vs OHCs: Differentiates IHC and OHC clusters with unique labels.
Directory Structure#
Outputs are saved in the Distances directory under the project folder. The Physical_distances.csv will have a new column called ‘CLass’ beside the ‘Distance’ column
Distances/
├── Physical_distances.csv # Includes updated cell type classifications.
Usage Instructions#
Perform clustering using the Perform Cell Clustering button.
Adjust or reassign cell types using the Re-assign Labels tool.
Review and validate the updated clustering in the viewer.
Save changes for downstream analysis.
Output Examples#
Clustered Cells: Visualizes cells with distinct colors:
IHC: Blue.
OHC1: Yellow.
OHC2: Green.
OHC3: Purple.
Physical_distances.csv:
Includes the ID and Class columns with updated classifications.
This feature streamlines cell type identification and classification, supporting advanced analysis of cochlear data.
Extending the Functionality#
To add or modify functionality, edit the following file:
identify_celltype_action.py
—
—