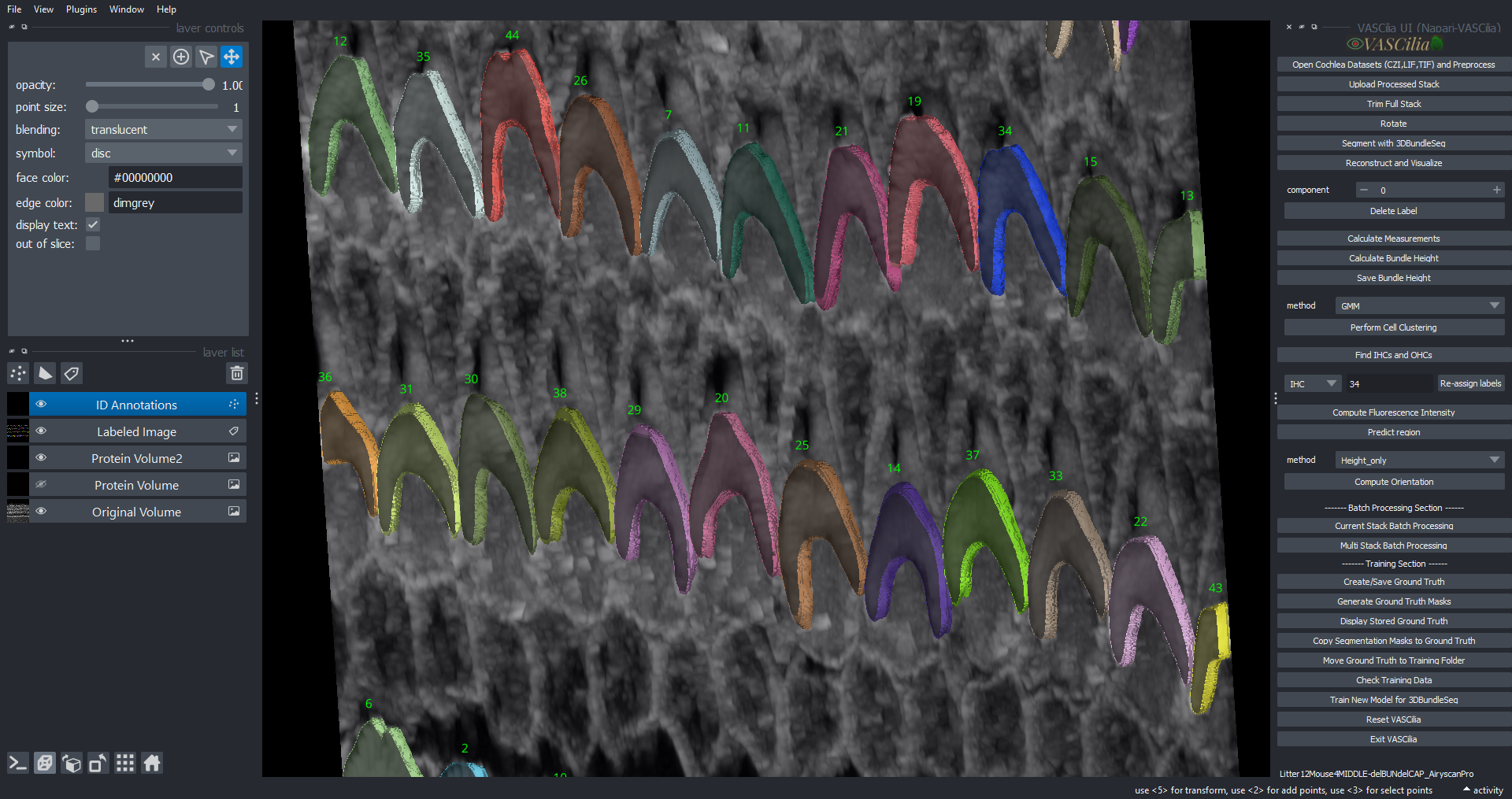
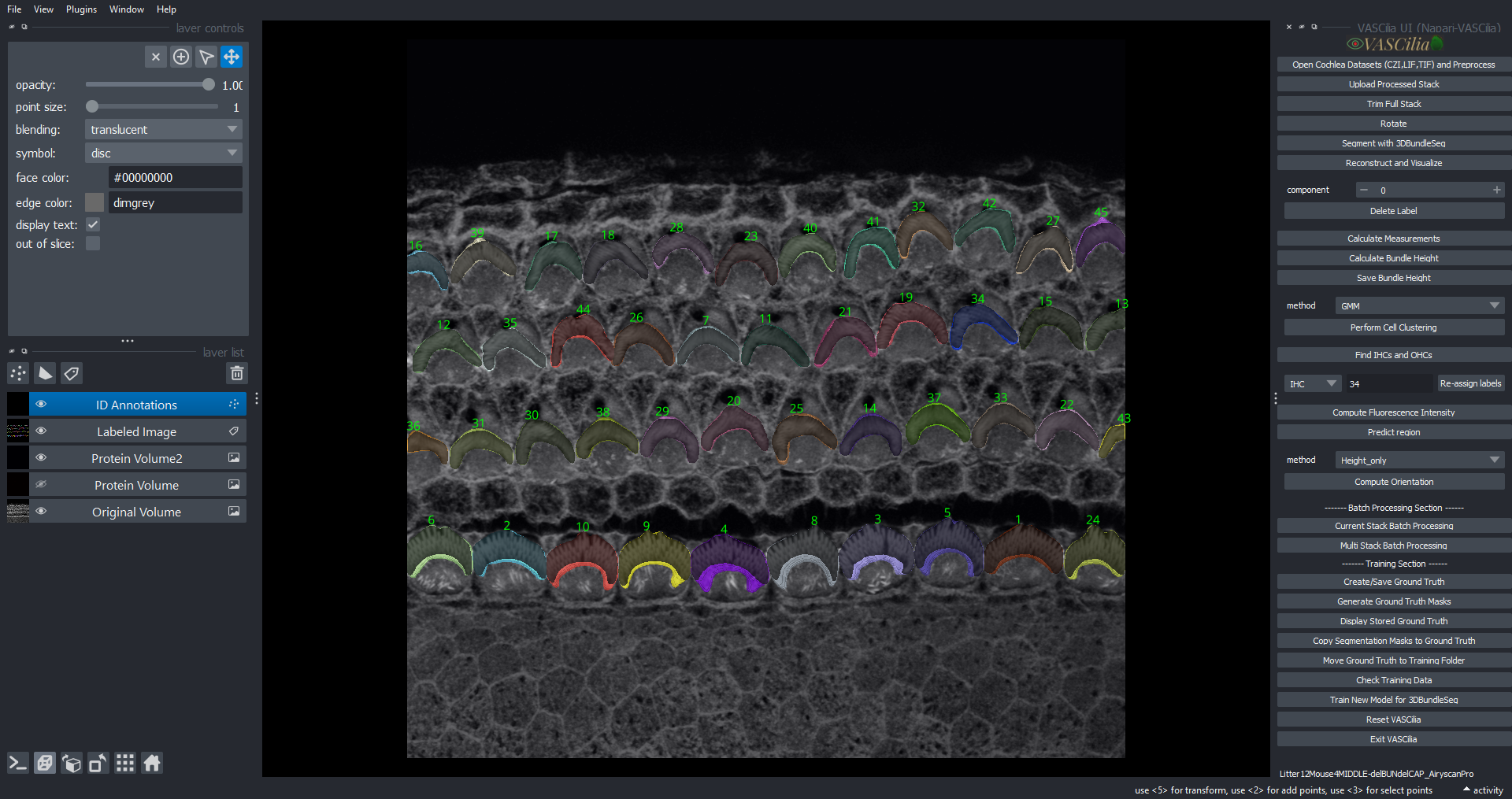
Visualizing and Tracking Cochlea Segmentations#
The Visualizing and Tracking feature in VASCilia facilitates the alignment and tracking of segmented cochlear regions across multiple image stacks to generate 3D reconstruction for the 3D object in which each 3D cell have one ID across all image frames. This tool ensures accurate identification and visualization of cellular structures, aiding downstream analysis.
—
Key Features#
### Overlap-Based Tracking
Tracks segmented components across frames by identifying overlaps with previous masks.
Assigns consistent labels to tracked components, ensuring continuity across the stack.
### 3D Volume Visualization
Combines individual frame segmentations into a single labeled 3D volume.
Provides detailed visualization of segmented components, including their properties and positions.
### Filtering and Refinement
Filters out regions with insufficient depth to improve tracking accuracy.
Updates the labeled volume to exclude filtered components.
### Annotation Support
Adds labels and annotations for segmented components directly in the Napari viewer.
Displays the highest point of each component, aiding in structural analysis.
—
Why Visualization and Tracking Are Important#
Cochlear stacks contain intricate cellular structures that require precise tracking to study their spatial and temporal changes. This feature:
Enhances understanding of cellular behavior over time.
Ensures accurate identification of regions across multiple frames for consistent analysis.
—
Batch Processing with SORT Algorithm#
For larger datasets, the SORT (Simple Online and Realtime Tracking) algorithm can be used for batch processing, enabling efficient and scalable tracking across numerous stacks.
—
Usage Instructions#
### Step 1: Start the Visualization Process
Click the Visualize and Track button in the plugin interface.
Ensure the segmentation step is completed before starting tracking.
### Step 2: Process the Segmentations
The plugin will:
Load .npy files containing segmentation masks and properties.
Generate labeled masks for each frame.
Track segmented components across frames based on overlaps.
### Step 3: View the Results
Visualize the tracked components in 3D using the Napari viewer.
Access detailed annotations and labels for each component, including their highest points.
—
Technical Details#
### Overlap Detection Tracks components across frames using an overlap-based method:
Identifies the label of the overlapping region in the previous frame.
Assigns consistent labels to maintain continuity across frames.
Example:
def overlap_with_previous(component, previous_mask):
overlap = np.bincount(previous_mask[component].flatten())
overlap[0] = 0 # Ignore the background
return overlap.argmax() if overlap.max() > 300 else 0
### 3D Volume Creation Combines labeled masks from individual frames into a 3D volume using:
`np.stack` to aggregate 2D masks.
`find_objects` to identify regions in the volume.
### Annotation Adds annotations to the Napari viewer for each labeled component:
Identifies the highest point of each component using:
highest_point_index = np.argmin(y_indices) x_highest = x_indices[highest_point_index] y_highest = y_indices[highest_point_index]
Displays labels and annotations in 3D for easy reference.
—
Practical Considerations#
Pre-Requirements:
Ensure the segmentation step is completed before initiating visualization and tracking.
Verify the .npy files are in the correct directory.
Filtering:
Components with insufficient depth are filtered out to improve visualization quality.
Batch Processing:
For large datasets, consider using the SORT algorithm for efficient batch tracking.
Output:
Tracked components are saved as .png files in the new_assignment_obj directory.
Annotations are displayed directly in the Napari viewer.
—
Extending the Functionality#
To modify or extend the tracking and visualization process, update the following files:
visualize_track_action.py: For tracking logic and visualization integration.
visualize_track_SORT.py: For SORT algorithm implementation and batch processing support.
—
—