3D Segmentation of Cochlea Stacks#
The 3D Segmentation feature in VASCilia leverages deep learning to segment cochlear hair cell stacks, providing precise identification of cellular regions for further analysis. This process is automated using a pretrained model and is seamlessly integrated into the workflow.
—
Key Features#
### Deep Learning-Based Segmentation
Utilizes a model trained on Detectron2, with the Mask R-CNN R-50 FPN 3x architecture.
Trained with 45 manually annotated cochlear stacks, ensuring high accuracy and reliability for segmentation.
Automatically segments 3D stacks to identify and analyze specific cellular regions.
### Integrated Workflow
Ensures segmentation occurs after the Rotation step, aligning the cochlear stacks with the PCP (planar cell polarity).
Updates the analysis stage upon successful segmentation, ensuring smooth progression through the pipeline.
### Customizable Parameters
Allows user configuration for:
Number of training iterations.
Threshold values for segmentation.
Output model paths for storing segmentation results.
—
Usage Instructions#
### Step 1: Prepare for Segmentation
Ensure that the stack is aligned using the Rotate functionality.
Verify that the analysis stage is set correctly; segmentation requires that the analysis stage is 3 (Rotated).
### Step 2: Execute Segmentation
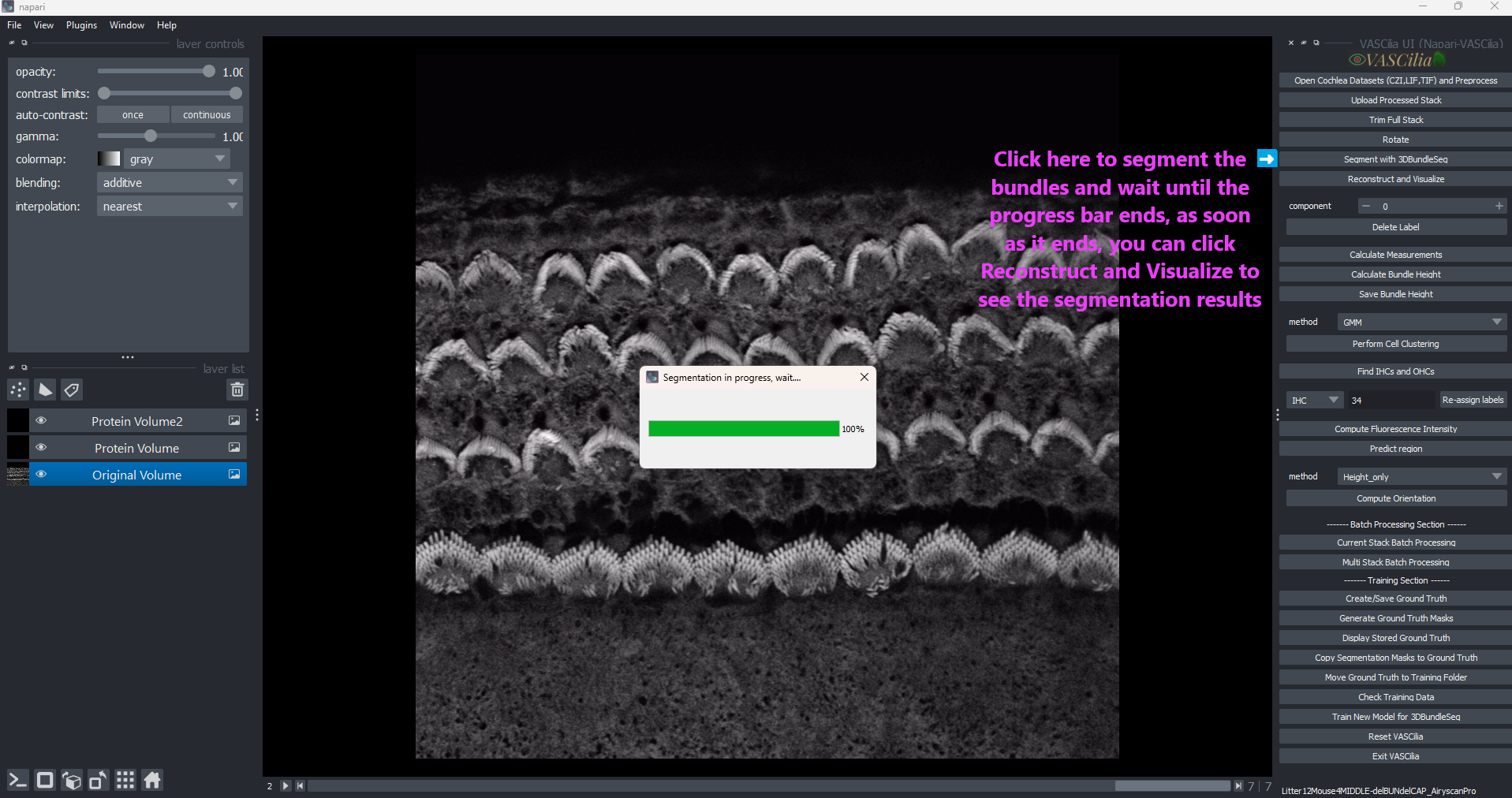
Click the Segment with 3DBundleSeg button in the plugin.
- The system will:
Prepare the dataset and paths.
Run the segmentation model using the configured parameters.
### Step 3: Monitor Progress
A progress dialog will display during segmentation, showing real-time updates.
Upon completion, results are saved in the designated output directory.
—
Technical Details#
### Pretrained Model
The segmentation model is trained using Detectron2 with:
Mask R-CNN R-50 FPN 3x architecture.
Trained with 45 manually annotated cochlear stacks to detect cellular regions with precision.
### Segmentation Command The plugin constructs and executes a command to perform segmentation, for example:
wsl {path_to_executable} --train_predict 1 \
--folder_path {train_folder} \
--model_output_path {output_model_path} \
--iterations {train_iter} \
--rootfolder {current_folder} \
--model {model_path} \
--threshold 0.7
### Progress Monitoring - The progress bar updates in real time by parsing output messages from the segmentation process. - The dialog closes automatically once the process completes.
—
Why Segmentation is Important#
Cochlear hair cells are intricate structures requiring detailed 3D analysis. Proper segmentation allows researchers to:
Identify cellular regions for focused study.
Quantify cellular dimensions and properties.
Ensure accurate downstream processing for visualization and analysis.
—
Practical Considerations#
Pre-Requirements:
Rotation of the stack is mandatory before segmentation.
Verify that the required wsl_executable path and model paths are correctly set in the config.json.
Error Handling:
If segmentation is attempted at the wrong stage, the plugin will notify the user to rotate the stack first.
If the stack has already been segmented, the plugin will prevent redundant processing.
Output Files:
The segmented results are stored in the output folder configured in config.json.
—
Extending the Functionality#
To add or modify functionality, edit the following files:
segment_cochlea_action.py: Handles the segmentation process.
To fine-tune our model with your images, use the Training Sction
—